About
This website allows you to estimate the most probable cancer cell line targets of a cytotoxic small molecule. The prediction is based on a combination of 2D and 3D similarity with a library of 80'000 known actives on more than 1000 cell lines.Developed and maintained by the Computer-aided molecular engineering group of the University of Lausanne (UNIL), Department of Oncology, Ludwig Institute for Cancer Research, and by the Molecular Modelling Group of the SIB Swiss Institute of Bioinformatics.
Ligand-based reverse virtual screening
The general pipeline of the SwissCellPrediction backend is summarized in the scheme below.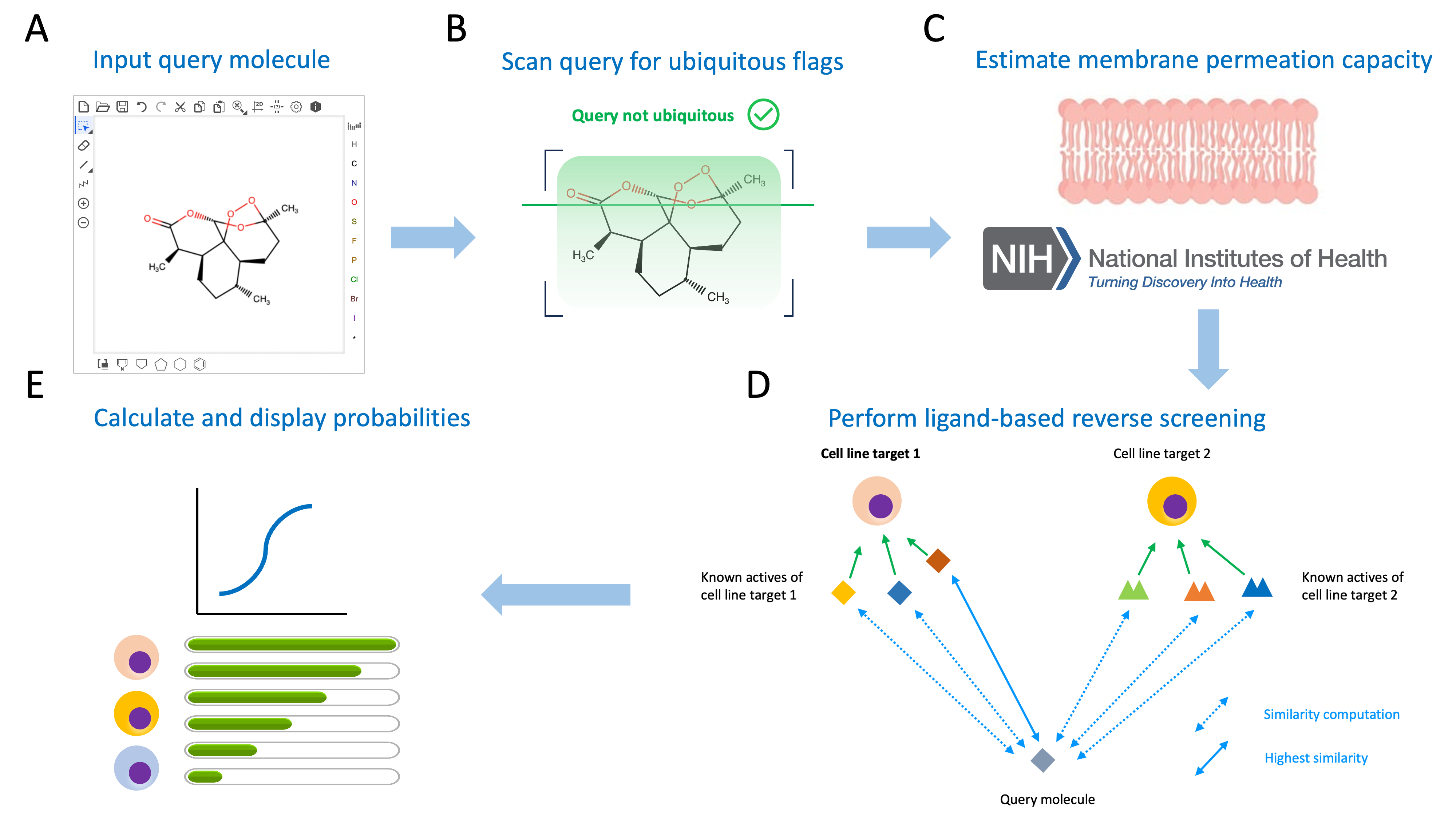 [A] Input of a query molecule by typing its SMILES or by using drawing or import options. [B] Substructure search inside query molecule for non-specific anticancer modes of action (i.e. ubiquitous compounds). In this example, the molecule (Artemisin) is not flagged as ubiquitous. [C] Estimation of the membrane permeation capacity of the query molecule using NIH PAMPA model. [D] Performing the reverse screening with pairwise similarity computations between the query molecule and every known active on each cell line target from the screening set. [E] Calculation of probabilities for each cell line to be a sensitive target of the query molecule, using the logistic regression model of SwissCellPrediction and display the results in a tabulated table.
[A] Input of a query molecule by typing its SMILES or by using drawing or import options. [B] Substructure search inside query molecule for non-specific anticancer modes of action (i.e. ubiquitous compounds). In this example, the molecule (Artemisin) is not flagged as ubiquitous. [C] Estimation of the membrane permeation capacity of the query molecule using NIH PAMPA model. [D] Performing the reverse screening with pairwise similarity computations between the query molecule and every known active on each cell line target from the screening set. [E] Calculation of probabilities for each cell line to be a sensitive target of the query molecule, using the logistic regression model of SwissCellPrediction and display the results in a tabulated table.
Case of ubiquitous query molecule
The general pipeline of the SwissCellPrediction backend in case of an ubiquitous query molecules is summarized in the scheme below. [A] Input of a query molecule by typing its SMILES or by using drawing or import options. [B] Substructure search inside query molecule for cytotoxic modes of action leading to non-specific anticancr agents (i.e. ubiquitous compounds). In this example, the molecule (Mitomycin C) is flagged as ubiquitous. [C] Estimation of the membrane permeation capacity of the query molecule using NIH PAMPA model. [D] Mapping the ubiquitous pattern found inside the query with the ubiquitous known actives of the cell lines available in the screening set of SwissCellPrediction, and display the results in a tabulated table.
[A] Input of a query molecule by typing its SMILES or by using drawing or import options. [B] Substructure search inside query molecule for cytotoxic modes of action leading to non-specific anticancr agents (i.e. ubiquitous compounds). In this example, the molecule (Mitomycin C) is flagged as ubiquitous. [C] Estimation of the membrane permeation capacity of the query molecule using NIH PAMPA model. [D] Mapping the ubiquitous pattern found inside the query with the ubiquitous known actives of the cell lines available in the screening set of SwissCellPrediction, and display the results in a tabulated table.







